FroggySection Data Adventure

This started when someone on the Graphics Programming discord shared a data archive from the Lawrence Berkeley Laboratories by the name of The Whole Frog Project, which a similar but much smaller scale operation when compared to what was performed during the preparation of the Visible Human Dataset. This process is called cryosectioning, and it consists of freezing a sample suspended in gelatin or other liquid, such that both subject and substrate are rigidly locked into place as a solid block. This is a common practice in biological sciences, and often times a lot of care must be taken not to damage the tissues while freezing. The intent of the data collected by this project was to construct a digital representation of the major organ systems of a frog, such that it might be used in place of frog dissection by students.
The Data
I think it's very interesting that I've found two public datasets which do very similar things, both of which being captured in the early 90s ( as an aside, if the reader knows of any others, I'd be very interested to know about them - please contact me with any details ). Parts of this data are all of 30 years old, at time of writing, and it's really quite remarkable to still be able to access and use it all these years layer. It's also quite interesting that there aren't many such datasets made publicly available in more recent years.
This type of data is very interesting to work with, and I'm finding that as the second dataset of this type that I've worked with, I feel I can comment more about it - namely, this cryosection setup was of much lower quality than the operation by the National Library of Medicine during the preparation of the Visible Human Dataset cryosections. The lighting on the slices is very inconsistent, and changes too much towards lower slices to have any level of consistent coloration to be used to segment the data in the way that I did the Visible Human. No color correction was done, and no color reference cards were included to do so.
Organ System Masks
To counter these shortcomings, and very unique to this dataset, organ system masks were manually prepared by the researchers in charge of the project. For each of the images of the cryosection slices, the major organ systems were traced out, by hand - per the Whole Frog Techincal Report:
After digitizing all the photos, the process of visual segmentation of the frog had to begin. Since the frog could only be seen in frozen cross section, identifying all the organs posed a challenge. Organs could only be seen as micron thin slices were cut away, causing them to look somewhat distorted. The visual segmentation process was also complicated by the fact that the representation of the organs had to be made with hand drawings using a computer mouse pointing device. The accuracy of the organs was thus limited to the experimentors drawing ability.
This would have been additionally complicated due to the inconsistent lighting conditions in the data. There is a notable absence in the form of the data for the skin - the data for the skin was unfortunately corrupted and unusable. The list of these masked organ systems include:
- Blood
- Brain
- Duodenum
- Retina of the Eye
- White of the Eye
- Interior of the Eyes
- Heart
- Ileum
- Intestine
- Kidneys
- Liver
- Lung
- Muscle
- Nerves
- Skeleton
- Spleen
- Stomach
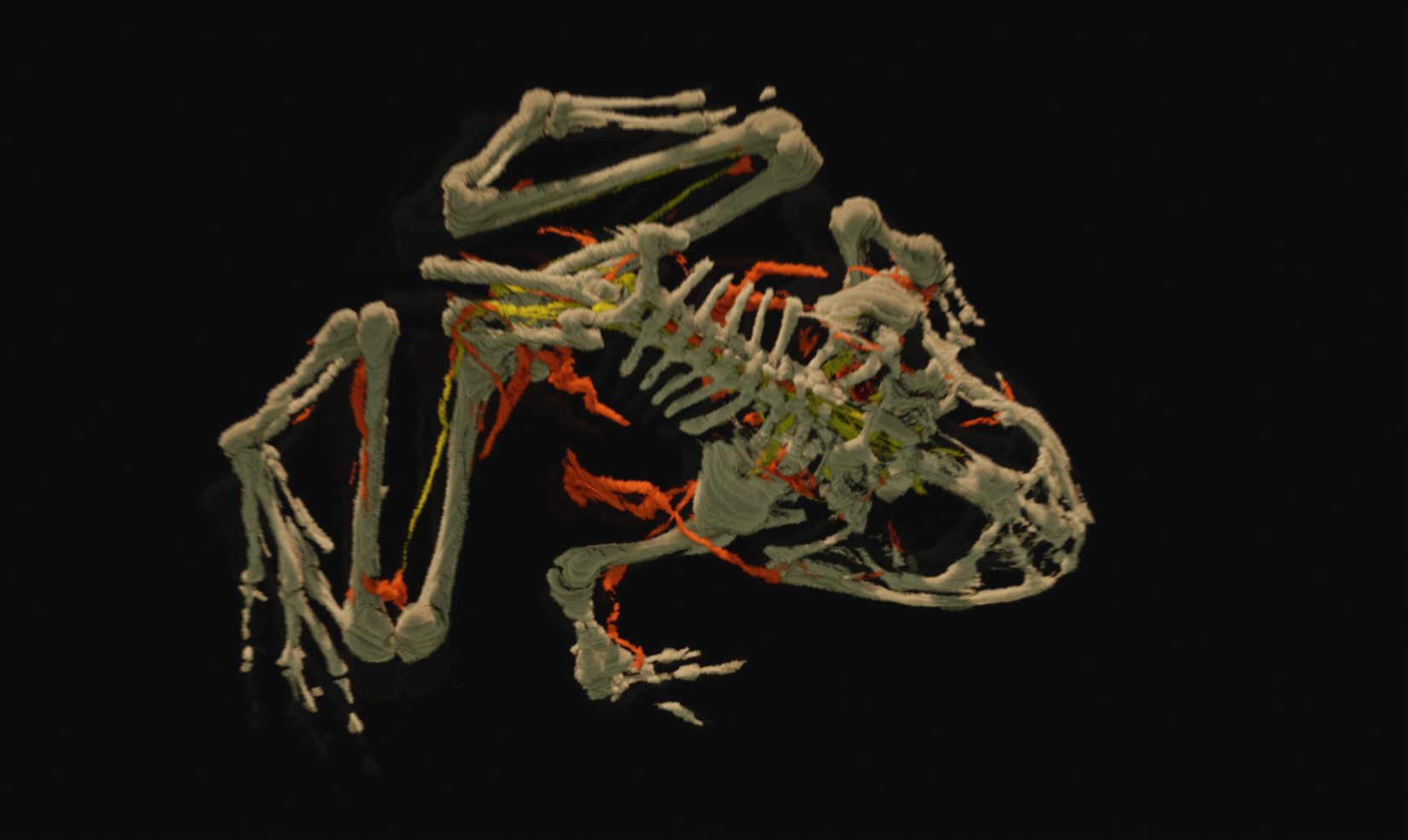
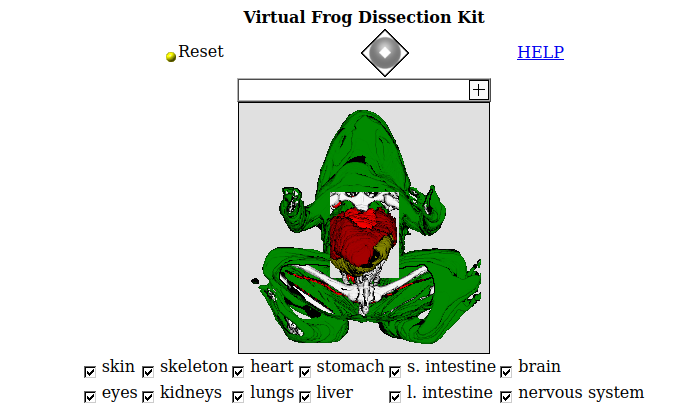
These masks are 1-bit images which indicate the regions occupied by each organ system. They are dimensionally self-consistent - by this, I mean that when overlaid they can be used to construct a false color voxel representation of the organ systems of the frog, several of which can be seen on this page, processed and lit in Voraldo 1.2. They are not, however, consistent with the original cryosection data, as they are scaled, offset and cropped - this adds a layer of complication if you wanted to try to get the color data lined up with the masks. In the original presentation of the data by Lawrence Berkeley Labs, they used a forms based applet called the Virtual Frog Dissection Kit, showing the top-down view of these systems you can see below, with a somewhat limited interface allowing visibilty for each system to be toggled by checkboxes. They have a little bit of information about how this 2d projection was prepared, again from the Whole Frog Technical Report:
Volume data is formed when all the slices of data images are stacked up. Each point in that volume space is called a voxel. A program named Sunvoxel is used to convert the 3D segmented image data of the frog into a projected view that can be displayed on the workstation screen. This program is capable of direct rendering of 3D data in a semi- transparent surface. It can handle multiple substances (e.g heart, lung, etc) with transparency and color value assigned to them. Each substance is classified by its range of voxel values. With the aid of the masks, the pixels of each substance of the frog are scaled to a certain range so that they can be treated as a distinct substance. With the classification, color, and opacity set, the frog can be rotated to any direction for viewing. By changing the opacity value of the substances, one can view objects that are blocked by other objects.
I can find no other reference to this Sunvoxel software in a cursory web search, it seems to be lost to history. It's an interesting preparation they have, with the outline contours of varying thickness to indicate the curvature of the various organs - it works well within the limitations that they had at their disposal. I think this same data could be used to construct a much higher fidelity frog dissection simulation with today's hardware capabilities, potentially even haptic simulation if links of material-appropriate properties could be formed between the neighboring voxels of each mask (rigid bones, etc).
Processed Data: FroggySection Repo
I have been working on preparing this data for use by others who might want to mess with it - there is accompanying MRI data for the frog, albeit at a much lower resolution, which I haven't messed with at all. I have the original cryosection data, the organ system masks, as well as the MRI data, all encoded as PNG. There's also a full mirror of the original froggy.lbl.gov site, acquired with wget. Shown below is all the organ systems except for the muscles, the voxels blurred slightly to deal with some of the aliasing issues present in the original slice data.
I think that a good thing to finish up the preparation of this data would be to pack the set of 17 masks into the bits of a PNG - since it's a lossless encoding, you've got 24 bits to work with - 8 each for red, green and blue. This leaves you 7 bits you could use to pack the MRI data into, if you were to upsample to a matching resolution and align it with the organ system masks. Depending on what the data contains, this would likely involve dropping one bit of precision from the MRI data. You might be able to mix two similar organ system masks ( eyes and eye whites, for example ) and get your 8 bit grayscale encoded in full.
This site also included similar datasets on some fruit and other animals. I have not looked closely at what is contained in this data, but it is all in the site mirror in the FroggySection repo.