Visualizing the Visible Human Dataset



This is an incredible resource which is made available by the National Library of Medicine. As specified on their website, they require that they be credited visibly whereever the data is used. I found a large FTP archive of the original data and some 4k rescans of the original film captures some time ago, and transferred the entire archive to local storage. There are several high resolution volume datasets, the largest of which is 4096x3051x5190 RGB voxels and 195 gigabytes uncompressed.
The main color data is the result of a process called cryosectioning. It is common in biological sciences, but it is incredibly unique for it to be done on humans. There were two full cadavers which were donated in 1994 and 1995, to be suspended in gelatin, frozen, and sectioned. While this process took place, pictures were taken of the cut face at each slice. You can read more about it here. These images, when interpreted as slices, constitute the contents of the volume dataset, as you can see below. There is no alpha channel, it is simply RGB captures directly from a visible light camera. Also included in the archive was a lot of radiological data that I have yet to even begin to get into.
The Data Itself
I have been sitting on this archive for a while, because I have not had the space to practially unpack it. At a total size of 460+ gigs, it's really impractical to do the processing on an SSD. I got a 5 terabyte external drive, copied it over, and unpacked it. Through the use of bash scripts, ImageMagick, and a basic c++ program to convert their raw byte format, I was able to convert the data to lossless PNGs. Because the different datasets are stored in different formats, it has to be done on a case-by-case basis, and it is still a work in progress. I have 2 datasets out of about 8 converted to a PNG format, both the 4k rescans of the Visible Male and Visible Female. This PNG format gives a huge savings on disk, both of these datasets were less than half as large, with all the contents intact. At some point it may make sense to learn more about compressed formats like DICOM.
Rescan Differences
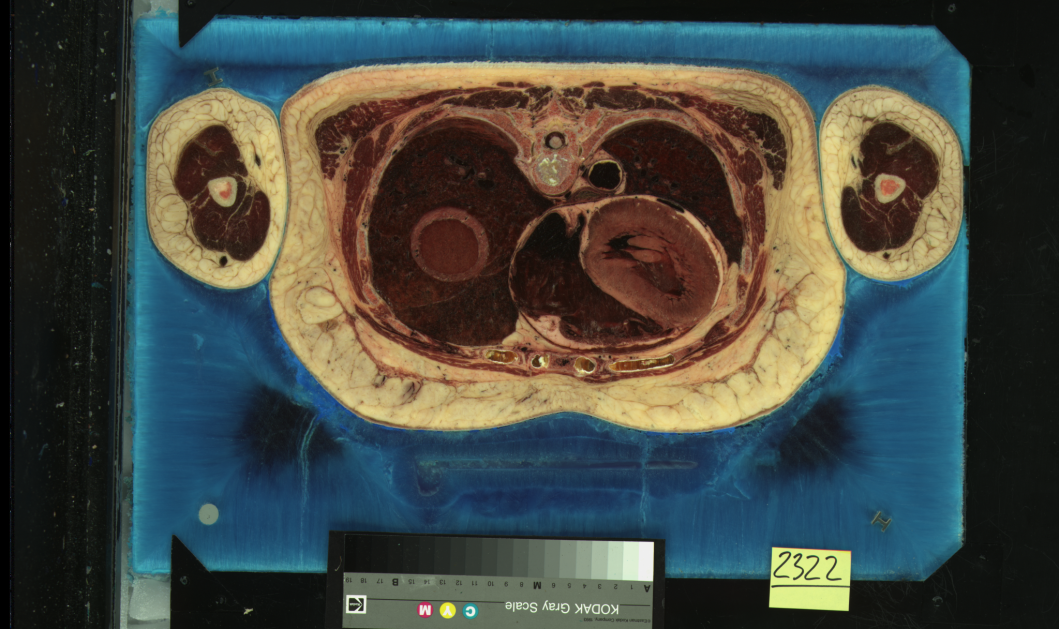
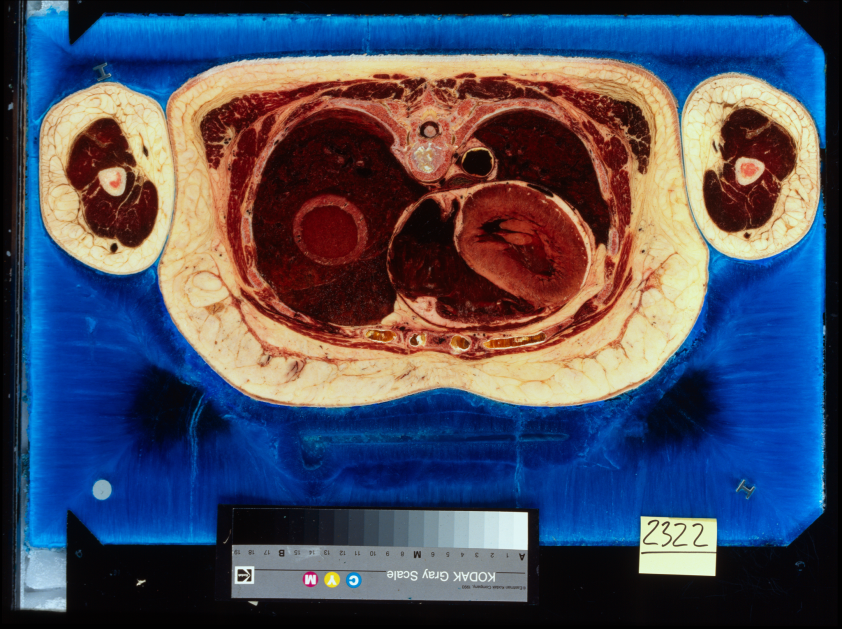
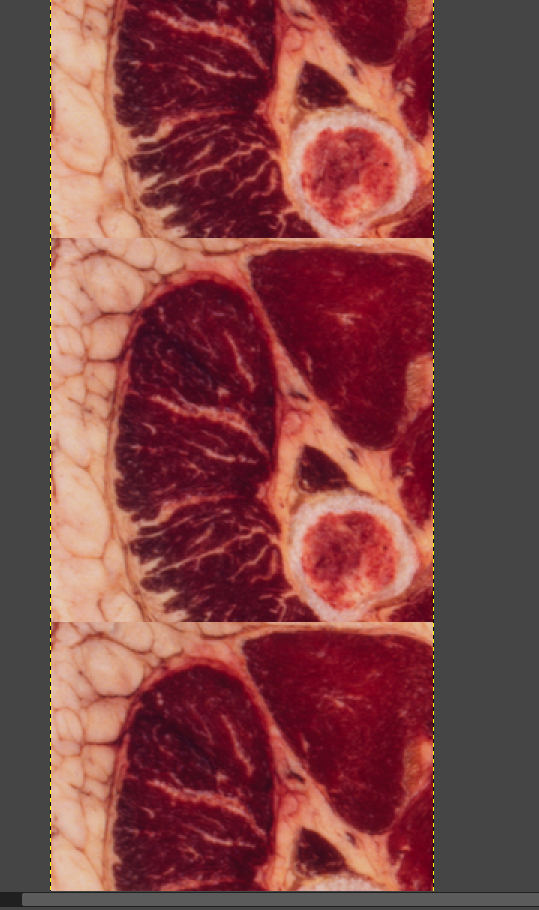
You can see the two versions of the same slice of the Visible Female dataset here. On the left is the original capture, and on the right is the 4k rescan, which was done on 10 year old film, in 2005. There are significant color and contrast differences, and my speculation is that the time is at least a factor. In the rescans, much of the fine detail seems blown out by comparison, with the colors shifted towards the blue end of things. I think that at least some of this was done to compensate for the green cast that existed in the original scans. In any event, the two datasets are quite distinct.
Segmenting the Data
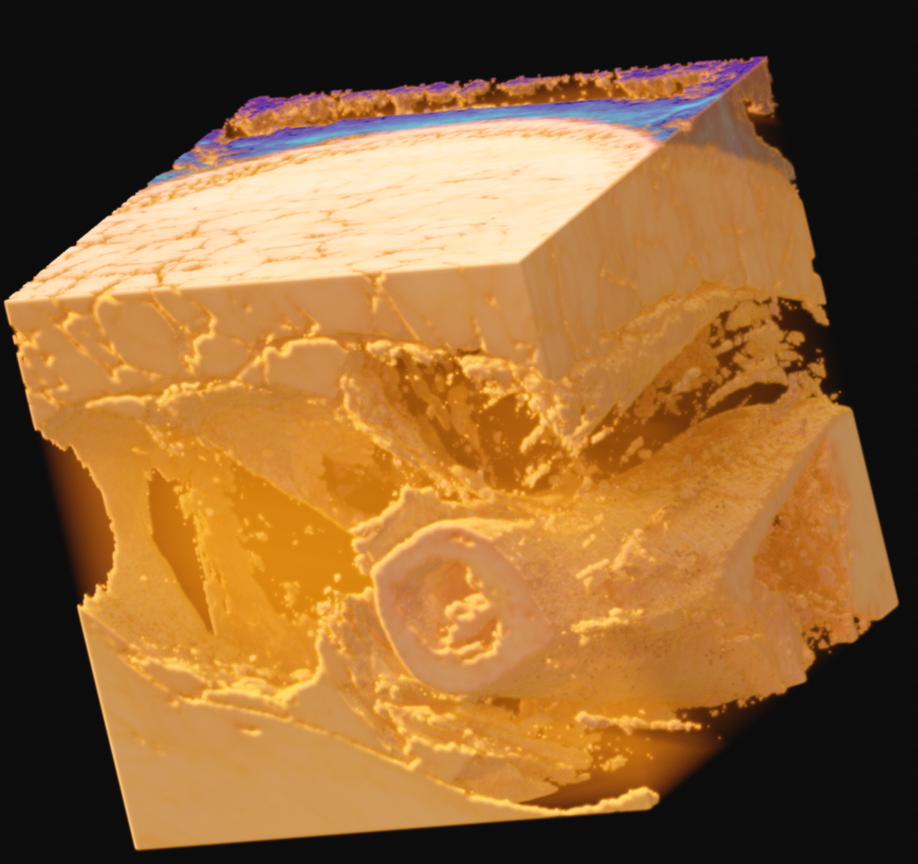
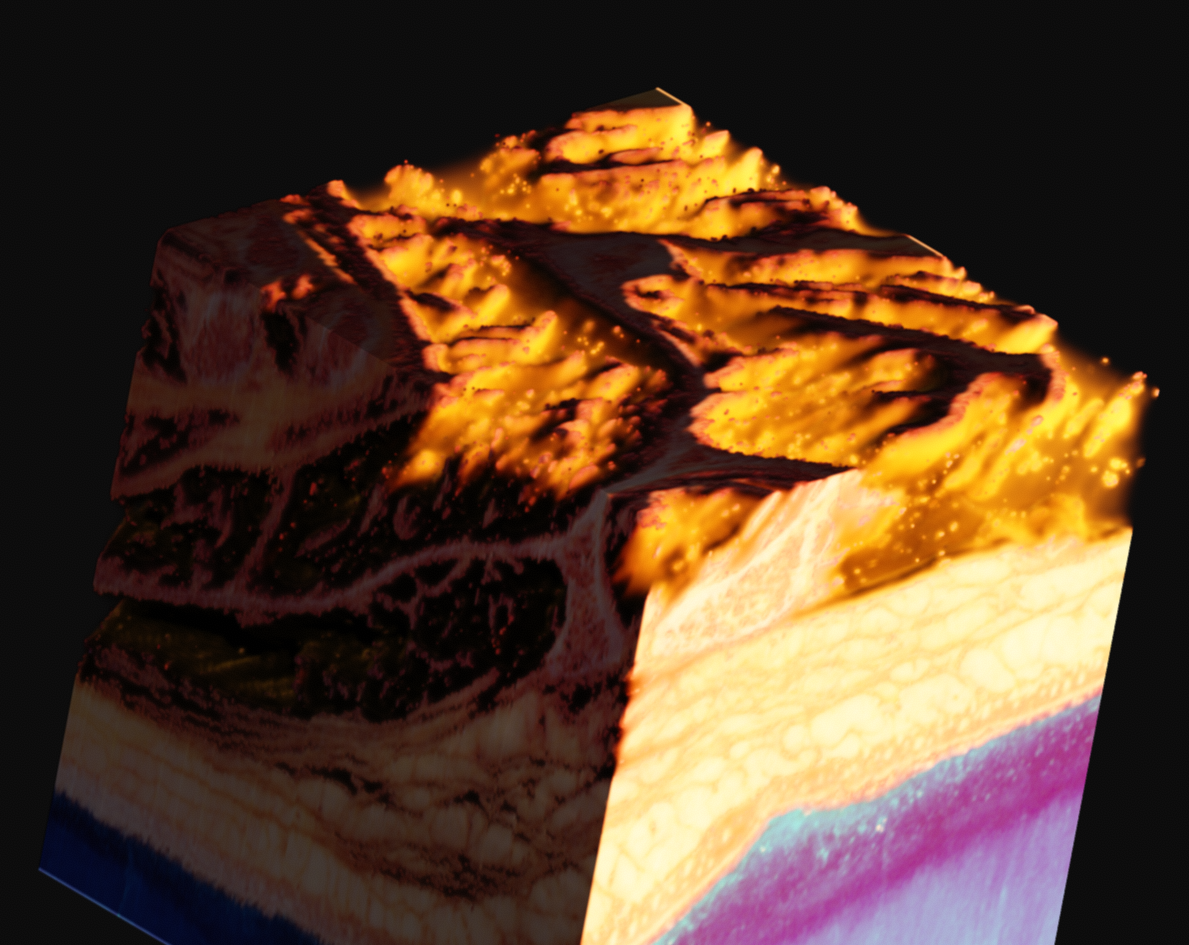
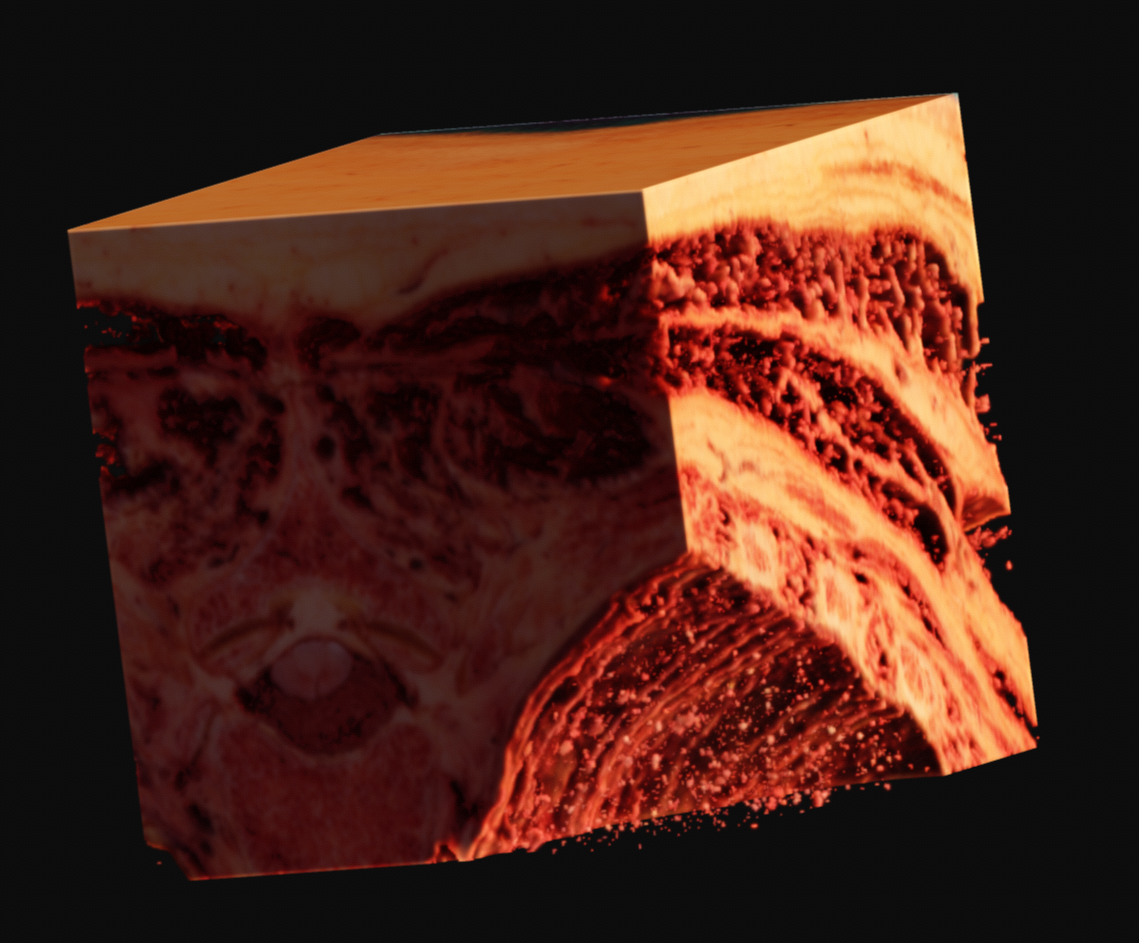
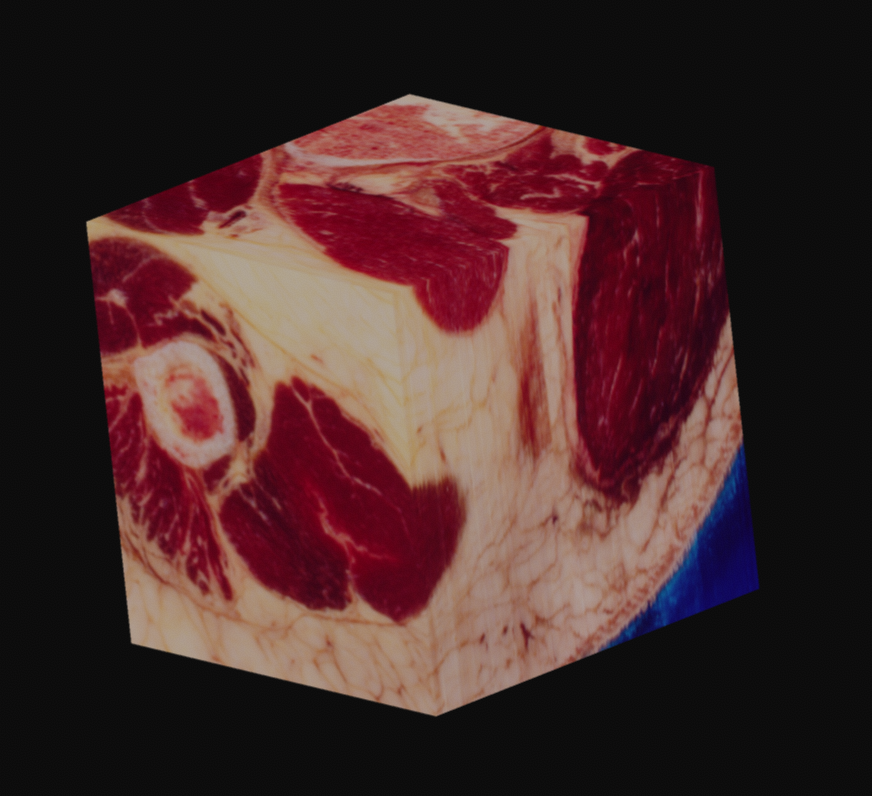
Since this data is provided as RGB, with no alpha channel, you are left to your own devices. In my case, I came up with what seemed like a relatively elegant solution using the existing tools in Voraldo. By masking the voxels which match certain criteria, specifically, values in the green channel greater than some threshold, I can segment the data to some degree. This use of the green channel is based on the idea that the read of the muscle has almost exclusively data in the red channel, the surrounding ice only has data in the blue channel, which leaves the green channel as an indpendent vector along which to classify. This caught white sections, such as bone, tendons, fat, and skin.
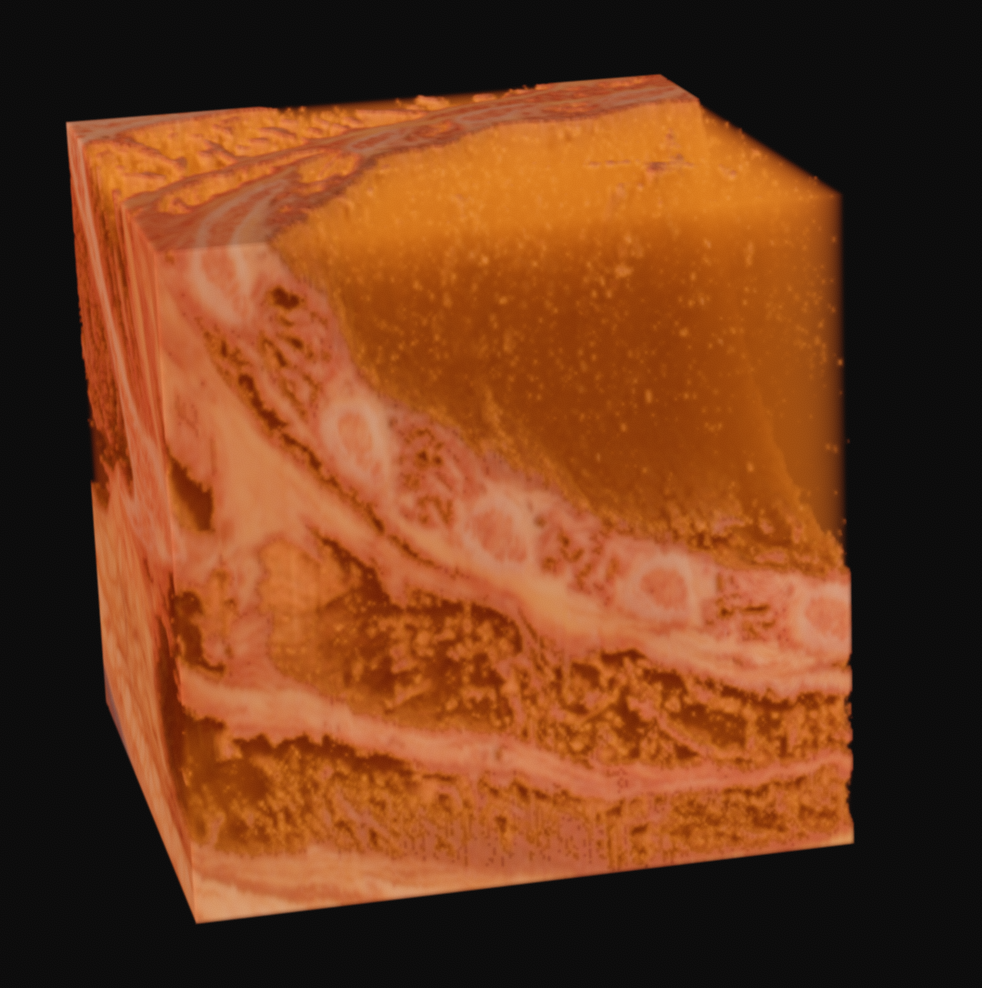


With variation on this logic, it's possible to segment just the muscle, or to isolate sections of the ice crystals. The data can be difficult to work with, as there is a lot of natural variation in the tissues. What you see in many of these images is a voxel gaussian blur applied, which respects the state of masked voxels. This gets rid of sharp edges by diffusing the color and alpha values out into the negative space.

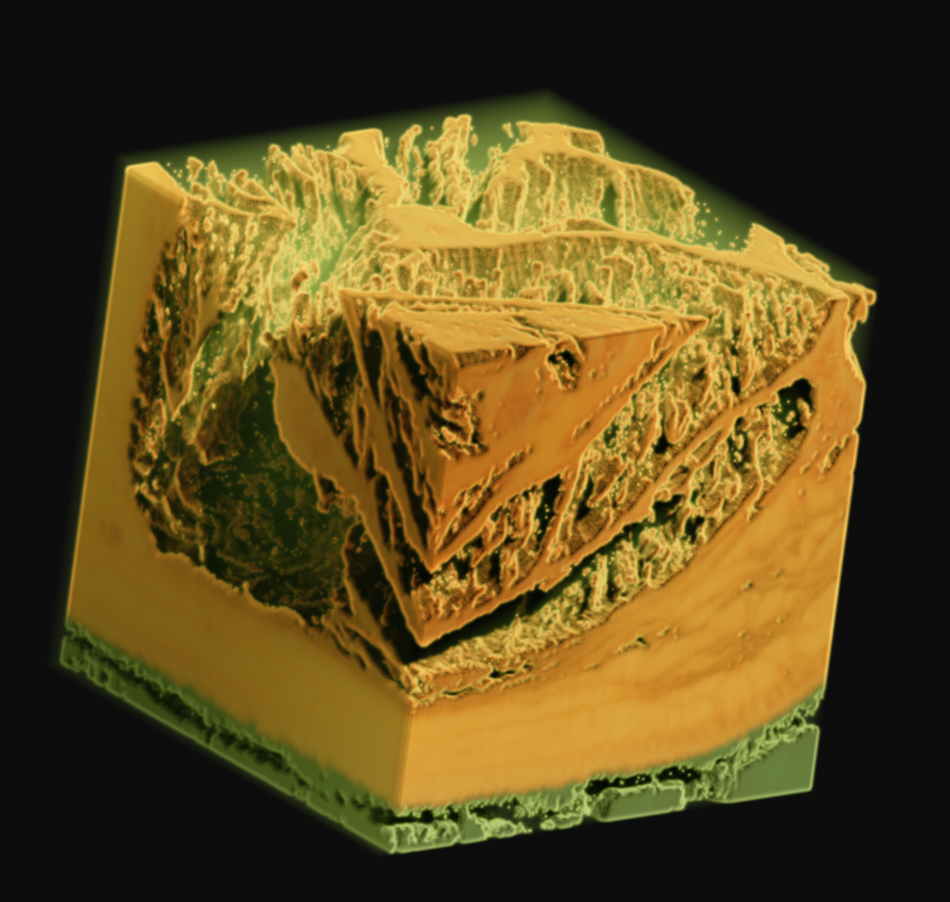

As one final image here, I have one rendered with the new perspective projection in Voraldo. You can see the slight foreshortening, and the DoF type of effect that comes from the ray origin jitter. There are still some slight issues with the uniform alpha value inside of the tissue, but I feel that this turned out fairly well, all things considered.
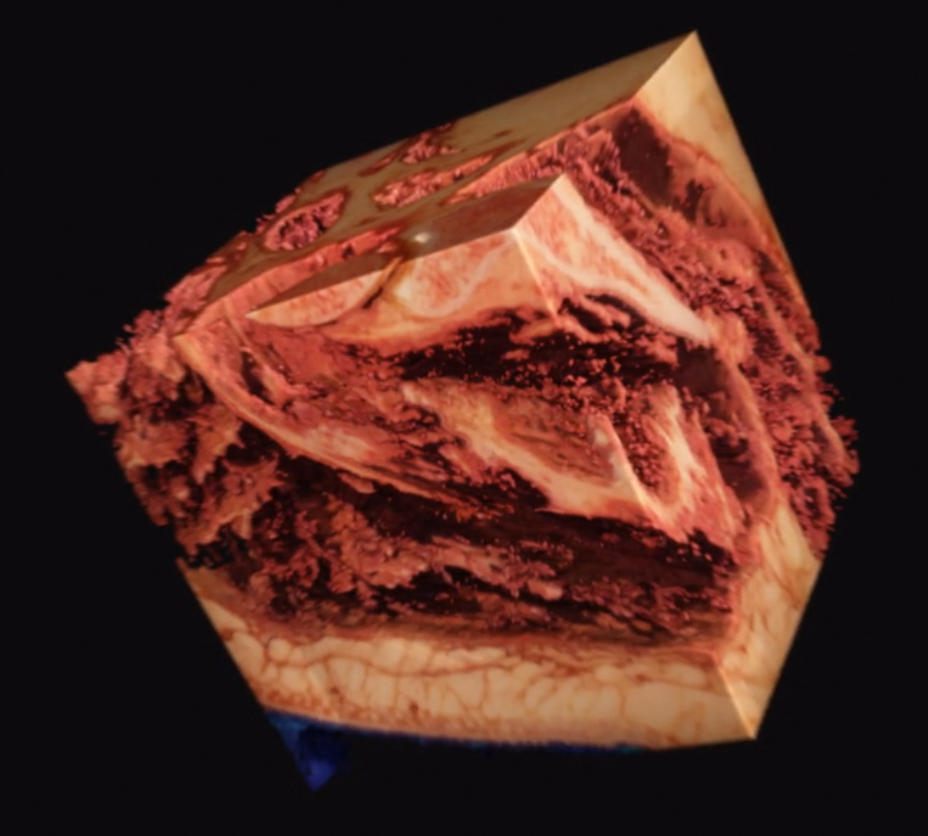
Future Directions
In the future, I may mess with this more, to try to segment each tissue as a distinct 'layer' of the data. I will also be generating a mip chain of each dataset and writing some code to resample arbitrarily scaled, oriented volumes out of the originals. I still need to finish processing copies of the originals to PNGs, as well.
I would also like to get into the radiological data, maybe try to line it up with the color data. There is also another project whose data is included, called the Visible Human 2 - this took finer sections of three heads, in coronal, saggital, and logitudinal sections, with more care taken to prevent damage to the tissues while freezing for cryosectioning. This data is very clean and should be interesting to work with, as there are pronounced blue and red veins and arteries which traverse the head and are visible. Masking off only these sections might give an interesting view of the vascularity of the head.